Introduction
This document contains information and examples on how to use the Lheuristic Shiny application to select L-shaped genes from two data matrices, an expression and a methylation matrix, matched by rows (genes) and columns(sample), using the algorithms implemented by the authors and described elsewhere.
The idea is to “keep-it-simple” so that it can serve as the basis for other applications such as a program that optimizes the parameters or for a graphical user interface based in Shiny.
Essentially what one user can do with this program is the following:
Program input
-
One can select demo data included in the web site or
-
One can upload its own data files
- A methylation data matrix (provided as a .csv or .txt file)
- An expression matrix (provided as a .csv or .txt file)
Both data matrices must have the same structure, that is same number of rows and columns with same names.
- Set parameter values
-
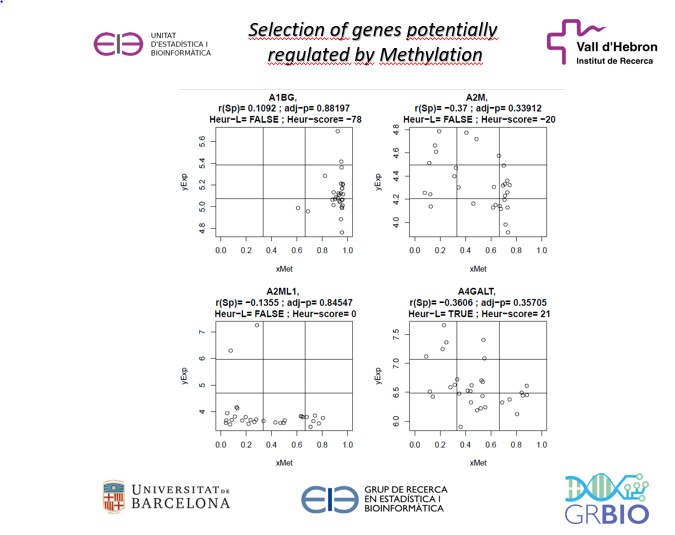
The computations performed to select L-shape genes require that certain parameters are set. These are described in more detail elsewhere but essentially they consist of:
- Number of genes in the data matrices to be analyzed
- Horizontal and vertical grid coordinates
- Minimum/maximum number of points in a cell for a gene to be L-shaped
- For cells: (1,1), (2,1), (3,1), (3,2) and (3,3) the number introduced is the “minimum” percentage of the minimum number points to be observed in an L-shaped gene
- For cells: (1,2), (1,3), (2,2), the number provided represents the “maximum” percentage of the total number of points to be observed in an L-shaped gene
- Weights given to each cell to compute the final score in genes that are declared “TRUE-L”
- Weights given to each cell to compute the final score in genes that are declared “FALSE-L”
Program output
Once the parameters have been set the application can be run and two different outputs are provided:
-
List of first selected and non-selected genes
-
(downloadable) .pdf file with names and scores of all genes
-
(downloadable) .pdf file with scatterplots for all genes
If you need more information or help about using the program ask met at: (asanchez AT ub DOT edu)